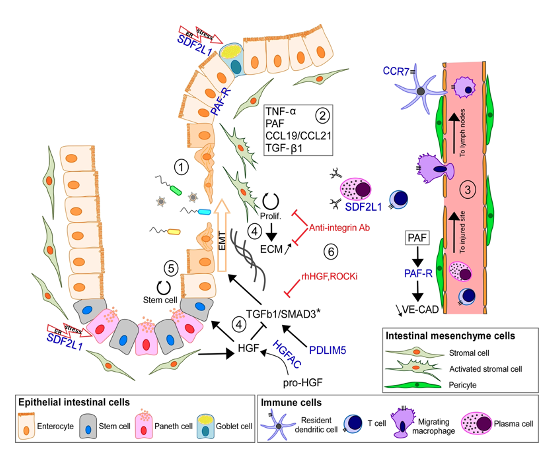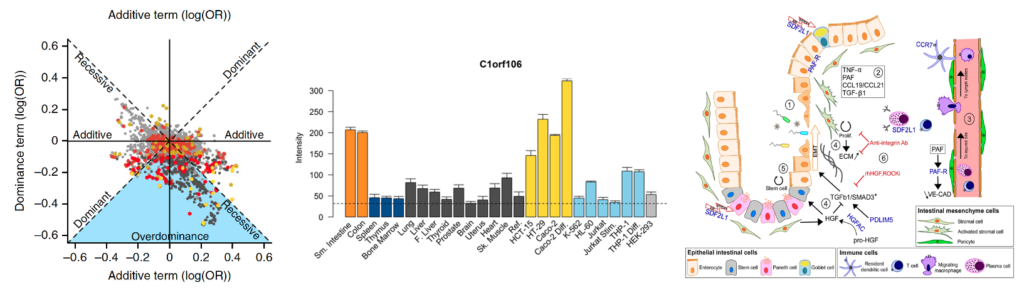
Using a combination of genome-wide association, fine mapping, DNA sequencing and transcriptomic studies we aim to identify causal genes, their protective and predisposing variants, and biological contexts in which they exert their influences. (images from PMID: 25559196, 29420262, 36038634).
We aim to define the rare and common variants, both coding and non-coding, that influence an individual’s probability of developing one of the inflammatory bowel diseases (IBD) known as Crohn’s disease (CD) and ulcerative colitis (UC).
It is important to identify protective and predisposing genetic variants for multiple reasons:
(1) to develop genetic tools such as polygenic risk scores that can aid in diagnosis;
(2) to increase success rates of drug development efforts; and
(3) to obtain a more comprehensive understanding of pathophysiologic mechanisms.
While some IBD genes may be ubiquitously expressed, many are enriched in specific cells and tissues. We therefore aim to identify the appropriate biological context in which to study IBD genes and their causal variants, for example the relevant tissues and cell types.
Selected Publications:
Large-scale sequencing identifies multiple genes and rare variants associated with Crohn’s disease susceptibility. Sazonovs A, et al. Nat Genet. 2022 Sep;54(9):1275-1283. doi: 10.1038/s41588-022-01156-2. Epub 2022 Aug 29.PMID: 36038634
Fine-mapping inflammatory bowel disease loci to single-variant resolution. Huang H, et al. Nature. 2017 Jul 13;547(7662):173-178. doi: 10.1038/nature22969. Epub 2017 Jun 28. PMID: 28658209
High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Goyette P, et al. Nat Genet. 2015 Feb;47(2):172-9. doi: 10.1038/ng.3176. Epub 2015 Jan 5. PMID: 25559196